Brownian Motion
Background
Brownian motion describes the stochastic movement of particles as they travel through space. This type of random movement is often referred to as a random walk, which is typical of particles that diffuse about a 1D 2D or 3D space filled with other particles or barriers. To understand Brownian motion, lets start by characterizing this phenomenon in 2D space.

The mathematical description of this process often includes these terms:
D (Δ): diffusion rate coefficient
D = L2 / 2d⋅t
L: step lengthd: dimensionst: time
D (in units: µm²/s)is the mean diffusion rate per unit time, often in µm²/s for biological motion on a molecular level. This refers to how fast, on average, the particle moves along its trajectories. This value is often of ultimate interest, particularly for simulating Brownian motion; however under most circumstances, the diffusion rate cannot be observed directly in empirical experiments - this would require the ability to visualize all the microscopic particles and collisions that dictate the particle's movement, and would have to be done on a nanosecond timescale. In the animation on the right, the diffusion rate can actually be quantified directly; but what is often seen when observing particle diffusion through a microscope would more closely resemble this: FIG: {{#info: {{{2}}} CLICK AWAY FROM IMAGE TO CLOSE }}. Instead, D is often calculated from the mean squared diffusion (MSD) path of the particle, defined below.
{{{2}}} CLICK AWAY FROM IMAGE TO CLOSE }}. Instead, D is often calculated from the mean squared diffusion (MSD) path of the particle, defined below.
MSD (Ω): mean squared displacement
MSD = 2d⋅D
MSD (in units: µm²)is the mean squared displacement of a particle over some time period.
K (κ): standard deviation of the normal distribution for D
k = √(d⋅D)
k (in units: µm)is the standard deviation (σ) of the normal distribution of step lengths that, when randomly sampled, will give rise to a diffusion rate D. This value is useful for simulating Brownian motion for a particular diffusion rate.
L (λ): mean step length
L = √(2d⋅D)
L (in units: µm)is the average step length per interval of observation. In diffusion simulations, this is the step size per iteration. This equation sometimes shows up in the equivalent formL = (2d * D).5
Lx (λˣ): mean 1D step length component of L
λˣ = L / √2
Lx or λˣ (in units: µm)is the average 1-dimensional step length for each component (X,Y,Z) dimension of L. For example, simulating 2D particle diffusion will require the generation of individual step lengths for both the X and Y dimension. The total step distance from the origin will be the length of the hypotenuse created by the individual X and Y component step lengths. In fact, the equation: λˣ = L / √2 is derived from the Pythagorean theorem for right triangles, such that 2(λˣ)² = λ² where 2(λˣ)² represents a² + b² and λ² represents c².
Ls (λˢ): step length scalar coefficient
Ls = 1 / √(D/Ds)
Ds: new desired diffusion rate
Ls or λˢ (in units: units)is a coefficient value that, when multiplied by each Lx component step length, will scale those lengths to achieve a new diffusion rate Ds. After scaling, the new diffusion rate = D/(D/Ds).
Brownian Motion Matlab Code
function [boom] = BradsBrownianMotionScript()
format compact;
format short;
close all;
% Note: This matlab script requires 'msdanalyzer' toolbox
%-------------############################------------------%
% STARTING PARAMETERS
%-------------############################------------------%
D = .3; % Diffusion Rate
Ds = .1; % Diffusion Scalar (Ds = Dn)
Dn = D/(D/Ds); % new D after scaling L
d = 2; % dimensions
dT = 1; % time step
k = sqrt(d*D); % stdev of D's step size distribution
MSD = 2*d*D; % mean squared displacement
L = sqrt(2*d*D); % average diagonal (2D) step size
Lx = L/sqrt(2); % average linear (1D) step size
Ls = 1/sqrt(D/Ds); % scales Lx values for Dn
MSDtest = [1 0 0]; % test: D, Dn, or L
Scale = 1/10; % scale of model
Ndots = 100;
Nsteps = Ndots;
xyl = ones(2,Ndots);
xyds = ones(2,Ndots);
lims = ((D+1)^2)*10;
%===========================================================%
% LIVE PARTICLE DIFFUSION
%-----------------------------------------------------------%
for t = 1:Nsteps
xyds = STEPxyds(Ndots, k);
[xyl] = AMPARSTEP(Ndots, xyds, xyl);
MAINPLOT(xyl, lims);
end
%===========================================================%
%===========================================================%
% MSD RANDOM STEPS ANALYSIS
%-----------------------------------------------------------%
tracks = cell(Ndots, 1);
stepN = 1;
for t = 1:Nsteps
xyds = STEPxyds(Ndots, k);
[xyl] = AMPARSTEP(Ndots, xyds, xyl);
[tracks] = MSDfun(stepN, Nsteps, tracks, xyds);
stepN = stepN+1;
end
MSDfunction(tracks,Ndots,Nsteps,D,Dn,L,dT,k,Scale,MSDtest);
%===========================================================%
% MSD UNIFORM STEPS ANALYSIS
%-----------------------------------------------------------%
stepN = 1;
for t = 1:Nsteps
xyds = stepsize(Ndots, Lx);
[xyl xyds] = MSDAMPARSTEP(Ndots, xyds, xyl, Ls);
[tracks] = MSDfun(stepN, Nsteps, tracks, xyds);
stepN = stepN+1;
end
MSDfunction(tracks,Ndots,Nsteps,D,Dn,L,dT,k,Scale,MSDtest);
boom = 'done';
%-------------############################------------------%
end % ## END MAIN FUNCTION ##
%-------------############################------------------%
%%
%%
%-------------############################------------------%
% ## SUBFUNCTIONS ##
%-------------############################------------------%
%-------------------------------------------%
% STEP SIZE GENERATOR
%-------------------------------------------%
function xyds = STEPxyds(Ndots, k)
xyds = (k * randn(2,Ndots));
end
%-------------------------------------------%
% MOVE PARTICLES MAIN FUNCTION
%-------------------------------------------%
function [xyl] = AMPARSTEP(Ndots, xyds, xyl)
for j = 1:Ndots
xyl(:,j) = xyl(:,j)+xyds(:,j);
end
end
%-------------------------------------------%
% LIVE DIFFUSION PLOT
%-------------------------------------------%
function [] = MAINPLOT(xyl, lims)
%-------------------------------------------%
xlim = [-lims lims];
ylim = [-lims lims];
zlim = [-5 5];
%=================================%
% MAIN 2D PLOT
%---------------------------------%
figure(1)
subplot(2,1,1),
AMPARPlot = gscatter(xyl(1,:),xyl(2,:));
axis([xlim, ylim]);
set(AMPARPlot,'marker','.','markersize',[6],'color',[1 0 0])
%=================================%
% 3D PLOT
%---------------------------------%
figure(1);
subplot(2,1,2),
gscatter(xyl(1,:),xyl(2,:)); view(20, 30);
axis normal;
grid off
axis([xlim, ylim, zlim]);
set(gca, 'Box', 'on');
end
%-------------------------------------------%
% MANUAL STEP SIZE FUNCTION
%-------------------------------------------%
function xyds = stepsize(Ndots, Lx)
%-------------------------------------------%
Lx(1:2,1:Ndots) = Lx;
xyd = randi([0 1],Ndots,2)';
xyd(xyd == 0) = -1;
xyds = (Lx.*xyd);
end
%-------------------------------------------%
% MSD SCALED STEPS FUNCTION
%-------------------------------------------%
function [xyl xyds] = MSDAMPARSTEP(Ndots, xyds, xyl, Ls)
for j = 1:Ndots
xyds(:,j) = xyds(:,j)*Ls;
xyl(:,j) = xyl(:,j)+xyds(:,j);
end
end
%-------------------------------------------%
% MSD TRACKS GENERATOR
%-------------------------------------------%
function [tracks] = MSDfun(stepN, Nsteps, tracks, xyds)
time = (0:Nsteps-1)';
xymsd = xyds';
xymsd = cumsum(xymsd,1);
tracks{stepN} = [time xymsd];
end
%-------------------------------------------%
% MSD TRACKS ANALYSIS
%-------------------------------------------%
function [] = MSDfunction(tracks,Ndots,Nsteps,D,Dn,L,dT,k,Scale,MSDtest)
SPACE_UNITS = 'µm';
TIME_UNITS = 's';
N_PARTICLES = Ndots;
N_TIME_STEPS = Nsteps;
N_DIM = 2;
oD = D; % raw µm^2/s
D = D*Scale; % to-scale µm^2/s
oDn = Dn; % raw µm^2/s
Dn = Dn*Scale; % to-scale µm^2/s
oL = L; % raw µm
L = L*Scale; % to-scale µm
dTbase = dT; % raw time-step
dT = dT*Scale; % to-scale time-step
k = k; % stdv of step distribution
ma = msdanalyzer(2, SPACE_UNITS, TIME_UNITS);
ma = ma.addAll(tracks);
disp(ma)
figure
ma.plotTracks;
ma.labelPlotTracks;
ma = ma.computeMSD;
ma.msd;
t = (0 : N_TIME_STEPS)' * dT;
[T1, T2] = meshgrid(t, t);
all_delays = unique( abs(T1 - T2) );
figure
ma.plotMSD;
cla
ma.plotMeanMSD(gca, true)
mmsd = ma.getMeanMSD;
t = mmsd(:,1);
x = mmsd(:,2);
dx = mmsd(:,3) ./ sqrt(mmsd(:,4));
errorbar(t, x, dx, 'k')
[fo, gof] = ma.fitMeanMSD;
plot(fo)
ma.labelPlotMSD;
legend off
ma = ma.fitMSD;
good_enough_fit = ma.lfit.r2fit > 0.8;
Dmean = mean( ma.lfit.a(good_enough_fit) ) / 2 / ma.n_dim;
Dstd = std( ma.lfit.a(good_enough_fit) ) / 2 / ma.n_dim;
Dheader1 = ['Raw Unscaled Values'];
Dhead1 = [' D Dn L'];
Ddat1 = [oD oDn oL];
disp(' ')
disp(Dheader1)
disp(Dhead1)
disp(Ddat1)
yourtesthead = ['YOU ARE TESTING DIFFUSION FOR:'];
if MSDtest(1)
yourtest = [' D: original diffusion rate'];
elseif MSDtest(2)
yourtest = [' Dn: new diffusion rate'];
elseif MSDtest(3)
yourtest = [' L: step length'];
else
yourtest = [' generic diffusion rate'];
end
disp(yourtesthead)
disp(yourtest)
disp(' ')
fprintf('Estimation of raw D coefficient from MSD:\n')
fprintf('D = %.3g ± %.3g (mean ± std, N = %d)\n', ...
Dmean, Dstd, sum(good_enough_fit));
% Retrieve instantaneous velocities, per track
trackV = ma.getVelocities;
% Pool track data together
TV = vertcat( trackV{:} );
% Velocities are returned in a N x (nDim+1) array: [ T Vx Vy ...]. So the
% velocity vector in 2D is:
V = TV(:, 2:3);
% Compute diffusion coefficient
varV = var(V);
mVarV = mean(varV); % Take the mean of the two estimates
Dest = mVarV / 2 * dT;
Dheader2 = ['Scaling to model...'];
Dhead2 = [' D Dn L'];
Ddat2 = [D Dn L];
disp(' ')
disp(Dheader2)
disp(Dhead2)
disp(Ddat2)
fprintf('Estimation from velocities histogram:\n')
fprintf('Tested D = %.3g %s, compare to scaled Des value of %.3g %s\n', ...
Dest, [SPACE_UNITS '²/' TIME_UNITS], D, [SPACE_UNITS '²/' TIME_UNITS]);
% printf('D.psd target value was %.3g %s\n', ...
% Dest, msdDpsd, [SPACE_UNITS '²/' TIME_UNITS]);
end
More About Brownian Motion
Anything below this line might not be up-to-date and/or consistent with the spotless masterpiece above.
MSD Explained
m = 2d*Dk = sqrt(d * D)λ = k * √2 / √πλ = (2d * D)0.5λᵃᵇ = √(2λ²)/2
- m is mean squared displacement
- d is dimensions (2)
- D is diffusion rate
- k is σ of sampling distribution that yields D (µ=0)
λ is mean of half normal distribution with σ = k- λ is the mean length of the XY step
- λᵃᵇ is the step size for both X and Y is λᵃᵇ
In statistical mechanics, the mean squared displacement (MSD or average squared displacement) is the most common measure of the spatial extent of random motion; one can think of MSD as the amount of the system "explored" by a random walker.
More Background
The Probability Density Function (PDF) for a particle in one dimension is found by solving the one-dimensional Diffusion equation. (This equation states that the position probability density diffuses out over time - this is the method used by Einstein to describe a Brownian particle.
As a particle travels, the molecule is jostled by collisions with other molecules which prevent it from following a straight line. If the path is examined in close detail, it will be seen to be a good approximation to a random walk. Mathematically, a random walk is a series of steps, one after another, where each step is taken in a completely random direction from the one before. This kind of path was famously analyzed by Albert Einstein in a study of Brownian motion and he showed that the mean square of the distance traveled by particle following a random walk is proportional to the time elapsed. In two dimensions this relationship can be written as:
r²=4•D•t
In 1D, since both forward and backward steps are equally probable, we come to the surprising conclusion that the probable distance travelled sums up to zero! This is clearly a useless property to calculate. If however, instead of adding the distance of each step we added the square of the distance, we realise that we will always be adding positive quantities to the total. In this case the sum will be some positive number, which grows larger with every step. This obviously gives a better idea about the distance (squared in this case) that a particle moves. If we assume each step happens at regular time intervals, we can easily see how the square distance grows with time, and Einstein showed that it grows linearly with time.
In a molecular system a molecule moves in three dimensions, but the same principle applies. Also, since we have many molecules to consider we can calculate a square displacement for all of them. The average square distance, taken over all molecules, gives us the mean square displacement. This is what makes the mean square displacement (or MSD for short) significant in science: through its relation to diffusion it is a measurable quantity, one which relates directly to the underlying motion of the molecules.
In molecular dynamics the MSD is easily calculated by adding the squares of the distance. The linear (i.e. straight line) dependence of the MSD plot is apparent. If the slope of this plot is taken, the diffusion coefficient D may be readily obtained.
At very short times however, the plot is not linear. This is because the the path a molecule takes will be an approximate straight line until it collides with its neighbour. Only when it starts the collision process will its path start to resemble a random walk. Until it makes that first collision, we may say it moves with approximately constant velocity, which means the distance it travels is proportional to time, and its MSD is therefore proportional to the time squared. Thus at very short time, the MSD resembles a parabola. This is of course a simplification - the collision between molecules is not like the collision between two pebbles, it is not instantaneous in space or time, but is `spread out' a little in both. This means that the behaviour of the MSD at short time is sometimes more complicated than this MSD plot shows.

Albert Einstein in a study of Brownian motion and he showed that the mean square of the distance traveled by particle following a random walk is proportional to the time elapsed. This relationship can be written as:
r²=2d•D•t
- r² MSD
- d dimensions
- D diffusion coefficient (diffusion rate)
- t time step
The next important equation is the standard deviation of the diffusion rate distribution. It would be more convenient to call this the step size distribution but the two values are not exactly proportional. That equation is given as:
k = sqrt(d * D * t)
- k σ or standard deviation of the diffusion rate distribution
- d dimensions
- D diffusion coefficent
- t time step
Knowing k we can then generate random steps from a normal distribution with a mean µ = 0 and sd σ = k pulling a value from this distribution for both the X and Y dimensions for each step. In Matlab this would look like:
xyds = k * randn(2,Ndots);
This will generate a random walk with a diffusion rate = D and MSD = 2d•D. We might also be interested in the average step size that is being generated from this distribution to create such a diffusion rate. This can be done by taking the mean absolute value of the generated steps:
mStep ≈ mean(mean(abs(xyds),2))
The analytical solution to this problem however is the mean µ of the half normal distribution, with a standard deviation σ = k :
λ = k * √2 / √π
mS = k*sqrt(2)/sqrt(pi)
However, this value cannot be used as the X and Y step size, if you're expecting to get back to D. What you need is the Pythagorean length of the XY combined step. Einstein solved this too:
λ = (4 * D)^.5;
mSt = sqrt((2*ld^2))/2;
I'm simulating Brownian motion. Each particle (dot) in the simulation take a random size step in a random direction. This is in 2D. Each iteration, a dot gets passed 2 values, a random step size for the X direction and Y direction. Voila! Brownian motion.
Now to characterize this motion. This wasn't an easy task, until this really smart dude came along and provided a few theorems. Albert Einstein showed that the mean square of the distance traveled by particle following a random walk is proportional to the time elapsed. This relationship can be written as (and hopefully the unicode looks like r^2 and not r2.. I won't put integers directly after variables):
- r² = 2d*D*t
- r² = mean squared displacement MSD (µm²)
- d = number of dimensions
- D = diffusion coefficient (µm²/s)
- t = time elapsed between each measurement (1 s)
Why is the mean squared displacement good to know? Well, creating Brownian motion is easy (like I mentioned above), but simulating Brownian motion based on real-life observations is not so easy. This is because we can't directly observe the step size (the distance traveled between particle collisions), but we can observe the MSD. Ultimately I'd like to estimate the average step size, but having a good estimate of the diffusion rate (D) would suffice for making a simulation based on real-life observations.
Say we're observing a particle through a microscope diffusing along a flat surface (d=2), and each second (t = 1) we write down its displacement (for both X and Y), squaring each value (**see footnote). When we're done, we can calculate the mean of these values and find that MSD=8 µm². We can now find D using the equation above.
- 8 µm² = (2*2) * D µm²/s * 1 s
- 8 µm²/s = 4 * D µm²/s
- 2 µm²/s = D µm²/s
Sweet, we have the diffusion rate. We can now use this value to make a simulation of our empirical observations.... But how? The simulation needs random step sizes, not a diffusion rate. Einstein figured this out too. He determined that the standard deviation (σ=k) of the distribution of random steps is:
k = sqrt(d * D * t)
From our observations:
k = sqrt(2*2*1) = 2
Now all we have to do is pull random values from a normal distribution with µ=0 σ=2 for each X and Y step size. If I do that and run a simulation - lets do 100 steps for 100 particles. boom:
Sweet, it worked! D = 2.0
We could stop there. But we're curious... What was the average step size it was creating to get that D? So we make a subroutine to track that information. We find that the average of the absolute values of the step sizes for each iteration are coming out one by one:
1.48, 1.61, 1.56, 1.51, 1.68
The average of all those values is around 1.59
Why? We can take a more analytical approach. Since we are using the absolute value of each step to calculate the mean step size, we are essentially sampling from a half normal distribution. The mean of a half normal distribution is:
µ = σ * sqrt(2) / sqrt(pi)
Our standard deviation was 2.
2*sqrt(2) / sqrt(pi) = 1.5958
Nice, that's pretty much what we got (1.59).
So, to recap, we started with a diffusion rate D=2, which, using Einstein's equation, helped us determine we needed to generate a bunch of steps from a distribution µ=0 σ=2 which resulted in a bunch of steps that were +/- 1.59 on average. Then as a check, we took the mean square of all 100 steps for each particle and calculated D which turned out to be 2.02 so everything worked as planned.
We could stop there, but will we? Of course not. Let's do one last check, to see if we completely understand what's going on. Say we don't know D or MSD. In fact, we don't have any real-life empirical information to go on. Perhaps all we want to do is generate some random walks, and if we're interested, we can use those walks to calculate the diffusion coefficient (D). Let's make 100 particles take 100 steps with a step size of 1.5958 (randomly negative or positive), aka the average step size from our experiment above.
Before we do this, any guess what D is gunna turn out to be?
D = 1.27
Wrong!!
Duh! This value cannot be used as the X and Y step size, if you're expecting to get back to D. What you need is the Pythagorean length of the XY combined step. Einstein solved this too. But first, a recap:
m = 2d*Dk = sqrt(d * D)λ = k * √2 / √πλ = (2d * D)0.5
- m is mean squared displacement
- d is dimensions (2)
- D is diffusion rate
- k is σ of sampling distribution that yields D (µ=0)
λ is mean of half normal distribution with σ = k- λ is the mean length of the XY step
See our new equation there: λ = (2d * D). Now, all we have to do to get the X and Y step length values, is to calculate what two values equals λ when using the Pythagorean theorem:
- a²+b²=λ²
since 'a' and 'b' can be the same value, lets just use:
- 2a² = λ²
and solve for 'a'
λᵃᵇ = √(2λ²)/2
so the step size for both X and Y is λᵃᵇ
Image Gallery
Notes
Finding the MSD of the simulation is (a trivial task) using a Matlab toolbox (see video below). The target extrasynaptic MSD was 0.1 µm²⁄s (from Choquet FIG: {{#info:  {{{2}}} CLICK AWAY FROM IMAGE TO CLOSE }}). Scaling the model PSD size and distance between synapses to real-world values based on the target MSD is not a trivial task, and may actually be related only indirectly. Just because a particle moves quickly, does not mean it moves from A to B quickly - it depends on how much stuff there is to collide with. Linear estimates have shown that glycine receptor movements along dendrites at a speed of 1–2 µm/min (.016-.008 µm/s) FIG: {{#info:
{{{2}}} CLICK AWAY FROM IMAGE TO CLOSE }}). Scaling the model PSD size and distance between synapses to real-world values based on the target MSD is not a trivial task, and may actually be related only indirectly. Just because a particle moves quickly, does not mean it moves from A to B quickly - it depends on how much stuff there is to collide with. Linear estimates have shown that glycine receptor movements along dendrites at a speed of 1–2 µm/min (.016-.008 µm/s) FIG: {{#info:  {{{2}}} CLICK AWAY FROM IMAGE TO CLOSE }}
{{{2}}} CLICK AWAY FROM IMAGE TO CLOSE }}
First, the randomly generated step-size was scaled to produce an MSD of 0.1 units²⁄step using the Matlab Brownian motion toolbox. Second, the dimensions of the model need to be scaled. It was found that an XY random step-size of µ=0.4 (σ=.2) units produced an MSE ≈ 0.1 units²⁄step. Next, the arbitrary 0.4 units need to be given meaning...
Given movements along dendrites at a speed of 1–2 µm/min (.016-.008 µm/s) observation, we can use a derivation of the MSD equation to help scale the model. The root-mean-square distance after N unit steps, with a step length of (L) is:
- d = L*sqrt(N)
In order to travel a distance d, N steps are required by this equation
- N = (d/L)²
- N = (1000 nm /400 nm)² = 6.25
- N = (2000 nm /400 nm)² = 25
- N = (4000 nm /400 nm)² = 100
- Lets say a particle is traveling linearly at a rate of 1 µm / 100 s
- And takes between 6-25 steps to cover 1-2 µm.
here are several good resources for explaining and calculating MSD:
VIDEO
{{{2}}}
MEAN SQUARED DISPLACEMENT
- Brownian Motion Mean Squared Displacement
- The goal of this calculation is to relate the simulated particle diffusion to real world values, namely velocity.
- Particle velocity will be a function of MSD x units ²⁄s which scales on space (units) and time (s) parameters.
- Space and time in the model are defined arbitrarily as Step_Size and Step where each Step a particle moves a distance randomly chosen from a normal distribution (µ=1,σ=.2)
- a step size of 1 unit/step will produce a brownian motion MSD of ~0.52 ±0.2 units ²/s
- empirical observations show that reasonable values for MSD are:
- PSD 0.01 µm ²/s
- synaptic 0.05 µm ²/s
- extrasynaptic 0.1 µm ²/s
- given an MSD of 0.52 ±0.2 units ²/s at the current parameters: 1 step = 1 unit (at µ=1,σ=.2), the model will need to be scaled such that particles move at an extrasynaptic rate of 0.1 µm ²/s.
- spines are on average 1 to 10 µm apart, if the model is comparing two spines 1 µm apart, they should be separated by 5 units of model space. This is because the current particle diffusion rate of the model is .5 µm ²/s and the empirical MSD is .1 µm ²/s
to make 0.1 units²⁄step ≈ 0.1 µm²⁄s. It was found that an XY random step-size of µ=0.4 (σ=.2) units produced an MSE ≈ 0.1 units²⁄step. Then, the arbitrary 0.5 units were given meaning (converted to 0.5 µm) by scaling the model according to real-world values (see below) by making 1 unit = 1 µm; as a convention, a subunit will be 1/10th of a unit, thus 1 subunit = 0.1 µm). The PSD areas were set to 3-subunits (.3 µm) square, 20 subunits (2 µm) apart, within a rectangular field 20 subunits (2 µm) wide and 60 subunits (6 µm) long. Given these scaled dimensions where 10 subunits ≈ 1 µm, a particle with an XY step-size of 0.5 units moving in a straight line, could theoretically go from PSD1 to PSD2 in 4 steps (obviously given the simulated particles are moving with Brownian motion, this lower-bound would be extremely rare).
Lab
All pages in this lab
II. Simulating Brownian Motion
IV. BMC Software
Introduction
This exercise shows how to simulate the motion of single and multiple particles in one and two dimensions using Matlab. You will discover some useful ways to visualize and analyze particle motion data, as well as learn the Matlab code to accomplish these tasks. Once you understand the simulations, you can tweak the code to simulate the actual experimental conditions you choose for your study of Brownian motion of synthetic beads. These simulations will generate the predictions you can test in your experiment.
In each section, Matlab code shown in the box to the left is used to generate the plot or analysis shown on the right. To use the code, copy it from the box on the left, launch the Matlab application, and paste the code into the Matlab Command Window. If you are new to Matlab, check out the Intro to Matlab page to help you get started.
Simulating Single Particle Trajectories
This exercise shows how to simulate the motion of a single particle in one and two dimensions .
One Dimensional Brownian Motion
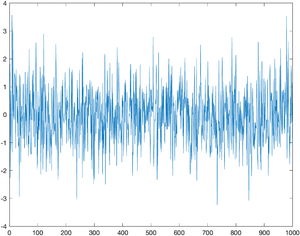
Brownian motion in one dimension is composed of a sequence of normally distributed random displacements. The randn function returns a matrix of a normally distributed random numbers with standard deviation 1. The two arguments specify the size of the matrix, which will be 1xN in the example below.
The first step in simulating this process is to generate a vector of random displacements. The commands to do this are shown below. N is the number of samples to generate.
N = 1000; displacement = randn(1,N); plot(displacement);
Distribution of Displacements
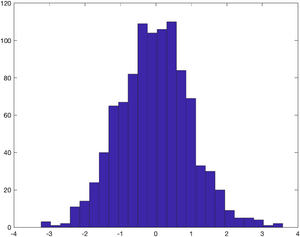
Have a look at the distribution of the randomly generated displacements. The hist command plots a histogram of the values. The second argument - 25 - specifies that Matlab should divide the values into 25 bins.
hist(displacement, 25);
Convert displacements to position
Now we have some appropriate random displacements. Their sum represents a particle trajectory in 1 dimension. The Matlab function cumsum returns the cumulative sum of a vector. The following commands take the cumulative sum of displacement and save the result in a vector called x.
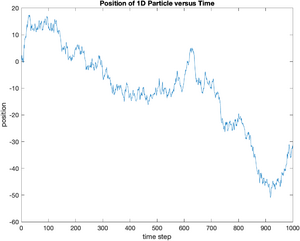
x = cumsum(displacement);
plot(x);
ylabel('position');
xlabel('time step');
title('Position of 1D Particle versus Time');
Two dimensional particle simulation
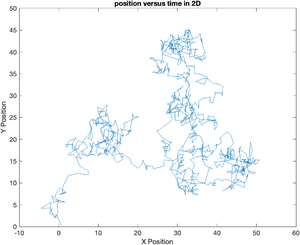
Extending this to two dimensions is simple. Since all directions are (assumed to be) equivalent, all we need to do is generate two vectors of random displacements. The vector of displacements saved in a Matlab structure called particle. x and y position vectors are stored in members of the structure. This data could also have been saved as a 2xN matrix. Using a structure has the important advantage that a meaningful name can be assigned to each member. This good practice makes your code much more readable.
particle = struct();
particle.x = cumsum( randn(N, 1) );
particle.y = cumsum( randn(N, 1) );
plot(particle.x, particle.y);
ylabel('Y Position');
xlabel('X Position');
title('position versus time in 2D');
Compute the Displacement Squared
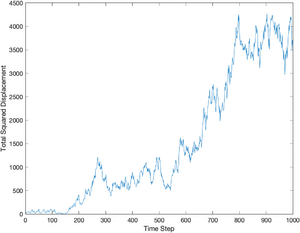
The displacement squared is equal to the x coordinate squared plus the y coordinate squared. Since the simulated particle always start at (0,0), it is unnecessary to subtract off the initial position (as will be necessary with the data you gather in the lab).
The dot caret (.^) operator raises each element of a matrix to a power.
dsquared = particle.x .^ 2 + particle.y .^ 2; plot(dsquared);
Theoretical Value of D
The theoretical value of the diffusion coefficient, D, is given by File:SingleParticleSimulations eq6021.png where T = temperature (Kelvin), kB = Boltzmann's constant, eta = viscosity, and d = radius.
Note that the units of D are length squared divided by time. See the lab writeup for more information. Let's compute D for a 1 micron particle in water at 293 degrees Kelvin.
d = 1.0e-6; % radius in meters
eta = 1.0e-3; % viscosity of water in SI units (Pascal-seconds) at 293 K
kB = 1.38e-23; % Boltzmann constant
T = 293; % Temperature in degrees Kelvin
D = kB * T / (3 * pi * eta * d)
ans =
4.2902e-013
A more realistic particle -- Getting the Units Right
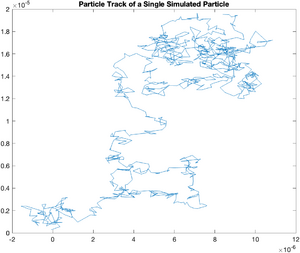
So far, we have been looking at simulated particles with a mean squared displacement of 1 unit per time interval. To accurately model a real particle, it is necessary to adjust the distribution of random displacements to match the experimental conditions.
According to theory, the mean squared displacement of the particle is proportional to the time interval, File:SingleParticleSimulations eq171153.png, where r(t) = position, d = number of dimensions, D = diffusion coefficient, and tau = time interval.
To generate the correct distribution, the output from randn (which has a standard normal distribution) must be scaled by the factor k.
dimensions = 2; % two dimensional simulation
tau = .1; % time interval in seconds
time = tau * 1:N; % create a time vector for plotting
k = sqrt(D * dimensions * tau);
dx = k * randn(N,1);
dy = k * randn(N,1);
x = cumsum(dx);
y = cumsum(dy);
dSquaredDisplacement = (dx .^ 2) + (dy .^ 2);
squaredDisplacement = ( x .^ 2) + ( y .^ 2);
plot(x,y);
title('Particle Track of a Single Simulated Particle');
Displacement Squared Plot
Theory predicts that the displacement should increase in proportion to the square root of time. The theoretical value of displacement squared is plotted with a thick black line. Since displacement is expected to increase with the square root of time, displacement squared is a straight line in the plot. With only a single particle and a small number of samples, deviation from the line can be quite large.
clf;
hold on;
plot(time, (0:1:(N-1)) * 2*k^2 , 'k', 'LineWidth', 3); % plot theoretical line
plot(time, squaredDisplacement);
hold off;
xlabel('Time');
ylabel('Displacement Squared');
title('Displacement Squared versus Time for 1 Particle in 2 Dimensions');
Estimating D from the Simulated Data
Solving for D gives File:SingleParticleSimulations eq250744.png. The best estimate of the value of D from the simulated data is:
simulatedD = mean( dSquaredDisplacement ) / ( 2 * dimensions * tau )
ans =
4.2192e-013
Uncertainty in the Estimate
The likely error of this measurement decreases as the square root of the number of samples. This will be discussed in more detail later.
standardError = std( dSquaredDisplacement ) / ( 2 * dimensions * tau * sqrt(N) )
actualError = D - simulatedD
standardError =
1.3162e-014
actualError =
7.1019e-015
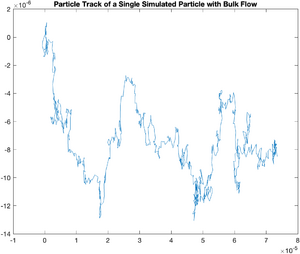
Systematic Error -- Bulk Flow in the Solvent
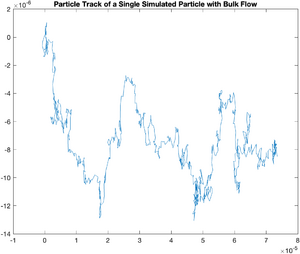
Sometimes, evaporation or uneven heating of the solvent will cause a flow to occur on the slide you are observing. We can model this easily. The following code models a flow with a magnitude 0.5 k in the x direction and 0.1 k in the y direction.
dx = dx + 0.2 * k;
dy = dy + 0.05 * k;
x = cumsum(dx);
y = cumsum(dy);
dSquaredDisplacement = (dx .^ 2) + (dy .^ 2);
squaredDisplacement = ( x .^ 2) + ( y .^ 2);
simulatedD = mean( dSquaredDisplacement ) / ( 2 * dimensions * tau )
standardError = std( dSquaredDisplacement ) / ( 2 * dimensions * tau * sqrt(N) )
actualError = D - simulatedD
plot(x,y);
title('Particle Track of a Single Simulated Particle with Bulk Flow');
simulatedD =
4.2926e-013
standardError =
1.3694e-014
actualError =
-2.3859e-016
Displacement Squared in the Presence of Bulk Flow
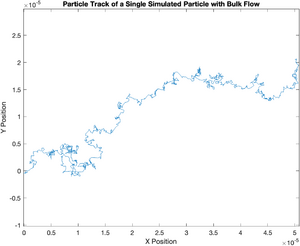
Notice how the plot of displacement squared diverges from the theoretical value. It has a distinct quadratic component. The magnitude of this error increases dramatically with time. This suggests that the error caused by bulk flow can be minimized by using the shortest possible sampling period. But there's a catch. As you increase the sampling rate, the amount of noise from the motion tracking algorithm goes up. A tenth of a second works pretty well for the particles you will observe in the lab. If you have time, take a few movies at a different rates to see the effect.
clf;
hold on;
plot(time, (0:1:(N-1)) * 2*k^2 , 'k', 'LineWidth', 3); % plot theoretical line
plot(time, squaredDisplacement);
hold off;
xlabel('Time');
ylabel('Displacement Squared');
title('Displacement Squared versus Time with Bulk Flow');
Simulating Multiple Particles
When you take your data in the lab, you will make movies of many particles. You will use a Matlab program to extract particle tracks from these movies. Because particles drift out of view and go in and out of focus, most movies will be about 5 seconds long at a sample rate of 10 Hz or so. Let's simulate this.
Using a For Loop to Generate Multiple Data Sets
A for loop is the key to generating multiple particle simulations. The results of the simulations are stored in a cellular array of structures called particle. For example, particle{3} refers to a structure containing the results of simulation number 3. particle{3}.D is the estimated value of D for simulation number 3. It is not necessary to understand this code in depth. But do yourself a favor and have a look.
particleCount = 10;
N = 50;
tau = .1;
time = 0:tau:(N-1) * tau;
particle = { }; % create an empty cell array to hold the results
for i = 1:particleCount
particle{i} = struct();
particle{i}.dx = k * randn(1,N);
particle{i}.x = cumsum(particle{i}.dx);
particle{i}.dy = k * randn(1,N);
particle{i}.y = cumsum(particle{i}.dy);
particle{i}.drsquared = particle{i}.dx .^2 + particle{i}.dy .^ 2;
particle{i}.rsquared = particle{i}.x .^ 2 + particle{i}.y .^ 2;
particle{i}.D = mean( particle{i}.drsquared ) / ( 2 * dimensions * tau );
particle{i}.standardError = std( particle{i}.drsquared ) / ( 2 * dimensions * tau * sqrt(N) );
end
SimulateParticle Function
That's a lot of typing. Fortunately, all of the commands to generate multiple particle tracks have been combined into in a single function called SimulateParticles. If you are interested in how the function is implemented, type edit SimulateParticles.m to have a look at the m-file for this function.
help SimulateParticles particle = SimulateParticles(N, particleCount, tau, k); usage: out = SimulateParticles( N, particleCount, tau, k ) N is the number of samples particleCount is the number of particles tau is the sample period k is the standard deviation of dx and dy returns a cellular array of length particleCount
Look at the Results
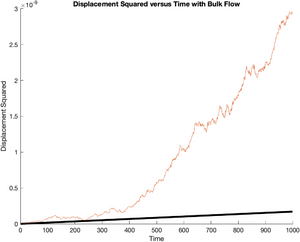
The following code plots all of the generated particle tracks on a single set of axes, each in a random color.
clf;
hold on;
for i = 1:particleCount
plot(particle{i}.x, particle{i}.y, 'color', rand(1,3));
end
xlabel('X position (m)');
ylabel('Y position (m)');
title('Combined Particle Tracks');
hold off;
Displacement Squared
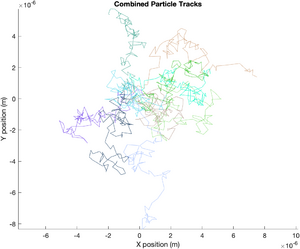
The following plot shows displacement squared versus time for all of the particles. The ensemble average of all the displacements is shown with a thick black line. The theoretical value of displacement squared is plotted with a thick blue line. Since displacement is expected to increase with the square root of time, displacement squared is a straight line in the plot.
% compute the ensemble average
rsquaredSum = zeros(1,N);
for i = 1:particleCount
rsquaredSum = rsquaredSum + particle{i}.rsquared;
end
ensembleAverage = rsquaredSum / particleCount;
% create the plot
clf;
hold on;
plot(time, (0:1:(N-1)) * 2*k^2 , 'b', 'LineWidth', 3); % plot theoretical line
plot(time, ensembleAverage , 'k', 'LineWidth', 3); % plot ensemble average
legend('Theoretical','Average','location','NorthWest');
for i = 1:particleCount
plot(time, particle{i}.rsquared, 'color', rand(1,3)); % plot each particle track
end
xlabel('Time (seconds)');
ylabel('Displacement Squared (m^2)');
title('Displacement Squared vs Time');
hold off;
Estimated Value of D
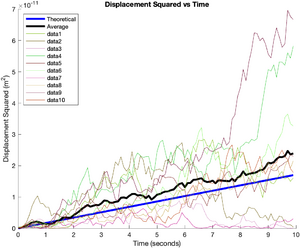
This plot shows the computed value of D for each simulation with error bars. A thick blue line indicates the best overall estimate of D (the average of the D value from each simulation) along with error bars in green * WHICH HAPPEN TO BE WRONG -- NEED TO FIX *.
clear D e dx;
% extract the D value from each simulation and place them all into a single
% matrix called 'D'
for i = 1:particleCount
D(i) = particle{i}.D;
dx(i,:) = particle{i}.dx;
e(i) = particle{i}.standardError;
end
% compute the estimate of D and the uncertainty
averageD = mean(D)
uncertainty = std(D)/sqrt(particleCount)
% plot everything
clf;
hold on;
plot(averageD * ones(1,particleCount), 'b', 'linewidth', 3); % plot estimated D
plot((averageD + uncertainty) * ones(1,particleCount), 'g-', 'linewidth', 1); % plot upper error bar
plot((averageD - uncertainty) * ones(1,particleCount), 'g-', 'linewidth', 1); % plot lower error bar
errorbar(D,e,'ro'); % plot D values with error bars
xlabel('Simulation Number');
ylabel('Estimated Diffusion Coefficient');
title('Estimated Diffusion Coefficient with Error Bars')
legend('Average Value of D', 'location', 'NorthWest');
hold off;
averageD =
4.2886e-013
uncertainty =
2.3294e-014
Fancy Statistics and Plots
This section looks at the statistical properties of the simulated data in more detail. In particular, it discusses:
- Uncertainty in the estimate of the mean value of a random variable from a population of samples
- The effect of squaring a normally distributed random variable
- The assumption of statistical independence of samples
A More Perfect Distribution


Did you notice that the distribution of random displacement values in the second section looked a little lopsided? Even with 1000 samples, there can be some noticeable deviations from the ideal distribution. A million samples looks quite a bit closer to the ideal.
hist(randn(1,1e3),25)
xlabel('Value');
ylabel('Frequency');
title('Histogram of Values for 1000 Samples of randn');
hist(randn(1,1e6),100)
xlabel('Value');
ylabel('Frequency');
title('Histogram of Values for 1000000 Samples of randn');
Sampling Uncertainty

This lab requires you to make an estimate of the average value of a random variable (actually, the square of a random variable) from a population of samples of that variable. The uncertainty in your estimate decreases with the square root of the number of samples, N. (That is, the standard error = 1/sqrt(N).)
This concept is illustrated in the following plot. To create the plot, fifty populations of N random samples are created for each value of N from 1 to 100. The mean value of each sample population is plotted with an 'x' versus N. The uncertainty appears to decrease as 1/sqrt(N) as expected. About two thirds of the values fall between the error bars (plotted in dark black).
Matlab note: check out the nested for loops used to create the plot.
clf;
hold on;
<span class="keyword">for i= 1:100
<span class="keyword">for j = 1:50
y = randn(1,i);
m = mean(y);
plot(i,m,'x');
<span class="keyword">end
<span class="keyword">end
plot(1:100, 1./sqrt(1:100), 'k', 'LineWidth', 2); % plot upper error bar in dark black
plot(1:100, -1./sqrt(1:100), 'k', 'LineWidth', 2); % plot lower error bar in dark black
hold off;
xlabel('Population Size (N)');
ylabel('Population Mean');
title('Population Mean of Normally Distributed Random Variable versus Population Size');
What Happens when you Square a Random Variable?

We did not estimate D in the simulations from the x and y displacements directly. Instead, we computed the mean squared value of dx and dy. What is the distribution of the resulting value? Adding the squares of two normally distributed random variables results in a chi-squared distribution (with two degrees of freedom) whose mean value equal to the sum of the variances of each variable. Here is what the chi-squared distribution looks like:
dx = randn(1,1e6);
dy = randn(1,1e6);
drSquared = dx .^ 2 + dy .^ 2;
mean(drSquared)
var(dx) + var(dy)
clf;
hist(drSquared,100);
title('Chi Squared Distribution (2 DOF), 1000000 Samples');
ans =
1.9982
ans =
1.9982
100 Years of BIO Lab Data in 1 Second
So does the uncertainty of the squared and summed behave as expected? The following plot is a simulation of 5000 data sets - 50 at each value of N just as above. (This is what a plot of all the data from students doing the BIO lab for the next hundred years might look like. Some lazy groups decided to use very low values of N.)
clf;
hold on;
for i= 1:100
for j = 1:50
dx = randn(1,i);
dy = randn(1,i);
m = mean( dx .^ 2 + dy .^ 2 );
plot(i,m,'x');
end
end
plot(1:100, 2 + 2./sqrt(1:100), 'k', 'LineWidth', 2); % plot upper error bar in dark black
plot(1:100, 2 - 2./sqrt(1:100), 'k', 'LineWidth', 2); % plot lower error bar in dark black
hold off;
xlabel('Population Size (N)');
ylabel('Population Mean');
title('Population Mean of Chi Squared (2 DOF) Distributed Random Variable versus Population Size');
Planning Your Time in the Lab
Sampling error sets a lower bound on the uncertainty in your estimate of the diffusion coefficient from experimental data. To plan your time in the lab, it will be important to understand how much data you should take for each experimental condition. Because the particles go out of focus and drift away from of the observation area, it can be difficult to make movies longer than about 5 seconds. At a frame rate of 10 frames per second, how many movies would you have to make to achieve a final uncertainty of 10%? 1%? .1%? What level of uncertainty will you try for in the lab?
Auto Correlation - A Closer Look at the Data

The simulations so far have assumed that we are in the overdamped limit, or the inertia-less regime. This assumption makes generating simulated trajectories very easy - all you have to do is add up a bunch of properly distributed random numbers. Of course, just because something is convenient doesn't mean it's true.
There are many statistical tests for independence and normality. (My personal favorite is the Kolmogorov-Smirnov test – but never before 5:00 PM.) Auto and cross correlation are a good place to start testing for independence. Autocorrelation looks for a relationship between a variable and its past or future values. Cross correlation looks for a relationship between two variables. Matlab has a built in function to compute auto and cross correlations called xcorr.
By construction, the simulated displacements are independent. The change in position at one time should exhibit no relationship at all to the change at any other time. This can be verified with xcorr. If x is a vector, xcorr(x) returns the correlation of x with itself for all possible offsets. The length of the resulting autocorrelation sequence is 2N+1. The middle value always stands out because x correlates perfectly with itself when there is no offset. Specifying the option 'coeff' causes the xcorr function to normalize this value to exactly 1 --perfect correlation.
Also notice that the autocorrelation sequence is symmetric about the origin.
clf;
c = xcorr(particle{1}.dx, 'coeff');
xaxis = (1-length(c))/2:1:(length(c)-1)/2;
plot(xaxis, c);
xlabel('Lag');
ylabel('Correlation');
title('Particle 1 x-axis Displacement Autocorrelation');
Cross Correlation
With two vector arguments, xcorr(x,y) returns a cross correlation matrix. The cross correlation can be used to test the relationship (or lack thereof) between one particle's trajectory and another's. If two particles move independently, the cross correlations should be very small. The code below computes the cross correlation between the x displacements of particle simulations number 1 and 2.
clf;
c = xcorr(particle{1}.dx, particle{2}.dx, 'coeff');
xaxis = (1-length(c))/2:1:(length(c)-1)/2;
plot(xaxis, c);
xlabel('Lag');
ylabel('Correlation');
Title('Particle 1, 2 x-axis Displacement Cross Correlation');
Matlab Tricks
Matlab can generate (and plot) an amazing amount of data. For example, the xcorr function can compute the auto and cross correlations of a large set of data values. If A is an MxN matrix, xcorr(A) returns a size 2*M-1 x N^2 matrix whose columns contain the cross-correlation sequences for all combinations of the columns of A. The football shape of the data is a result of the way that the data is windowed when computing the correlation.
% create an array whose columns contain the dx values for each particle
for i = 1:particleCount
allDx(:,i) = particle{i}.dx';
end
% compute all possible auto and cross correlations
c = xcorr(allDx, 'coeff');
% plot the results
clf;
hold on;
for i=1:size(c,1)
plot(xaxis, c(:,i),'color',rand(1,3));
end
hold off;
xlabel('Lag');
ylabel('Correlation Coefficient');
title('All Possible Auto and Cross Correlations in the x Dimension');
So what does the autocorrelation sequence look like for a trajectory that is not generated from independent samples? In the following example, the displacement at each time interval depends in part on the displacement in the last interval plus a random number. The autocorrelation is not nearly as sharp as the one generated by independent random samples.
x = zeros(1,N);
c = 0.80; % degree of correlation; 0 to 1
step = randn(1,N);
x(2) = randn();
for t=2:N
x(t) = (c * x(t-1)) + ((1-c)*step(t));
end;
clf;
plot(xaxis, xcorr(x, 'coeff'));
xlabel('Lag');
ylabel('Correlation Coefficient');
title('Autocorrelation');
Articles
Michalet • 2010 • Phys Rev E Stat Nonlin Soft Matter Phys - PDF
Expand to view experiment summary
We examine the capability of mean square displacement analysis to extract reliable values of the diffusion coefficient D of single particle undergoing Brownian motion in an isotropic medium in the presence of localization uncertainty. The theoretical results, supported by simulations, show that a simple unweighted least square fit of the MSD curve can provide the best estimate of D provided an optimal number of MSD points is used for the fit. We discuss the practical implications of these results for data analysis in single-particle tracking experiments.
Czondor Choquet • 2012 • PNAS - PDF
Expand to view experiment summary
FIG: {{#info: Error creating thumbnail: File missing(A) Schematic diagramof themodel. Kinetic parameters include Dout (extrasynaptic diffusion), Din (synaptic diffusion), DPSD (diffusion coefficient of the PSD), kon (AMPAR/ scaffold binding rate), koff (AMPAR/scaffold dissociation rate), and kendo (endocytosis rate). (B) Simulated trajectory (50 s). The synapse is in green, the PSD in red, and dendrite borders in gray. Geometric parameters are: a (synapse spacing), b (synapse width), c (PSD width), w (dendrite width). (C) High-magnification 30-s trajectory of an AMPAR-bound Qdot on the surface of a DIV 9 neuron. and simulated (plain curves) AMPAR median diffusion coefficients obtained at different neuronal ages (DIV 4–15) or varying synaptic spacing (0.75–30 μm), respectively, were plotted against synapse density. The simulated curves were computed for different kon values, keeping koff: 0.1 s−1. (F) Interquartile distributions of AMPAR diffusion coefficients from experiments (black) and simulations (green). Neuroligin-1 expression, which doubled the number of Homer1c-positive puncta, was mimicked by a decrease in synapse spacing (a: 1 μm). Overexpressing PSD-95 was modeled by enhancing kon (2.5 s−1) to mimic an increase in PSD binding sites, plus an increase in PSD size (c: 0.4 μm). CLICK AWAY FROM IMAGE TO CLOSE }}
- Dendrite
- length: 10 - 60 µm
- width: 2 µm
- D.out: 0.05 - 0.3 µm²/s
- PSD
- length: 0.3 µm
- width: 0.3 µm
- distance: 1 - 30 µm apart
- PSA (perisynaptic area PSD pad)
- length: 0.3 µm
- width: 0.3 µm
- D.in: 0.03 µm²/s
Total synaptic area: .6 x .6 µm In some cases, we varied the size of the synapse and the PSD, keeping a constant ratio of 2 between them (b: 0.1–1.6 μm; c: 0.05–0.8 μm).
AMPARs bound to the PSD were allowed to diffuse at a very low diffusion coefficient DPSD: 0.0002 μm2/s, reflecting the intrinsic movement or morphing of the PSD. We neglected the turnover of individual scaffold molecules, typically much longer than the AMPAR diffusion time scale





